Research
My research applies theoretical and computational approaches to phylogenomics, the study of evolutionary relationships among species using genome-scale datasets. Within this broad theme, I have three primary research aims:
Detecting and characterizing introgression using genomic data
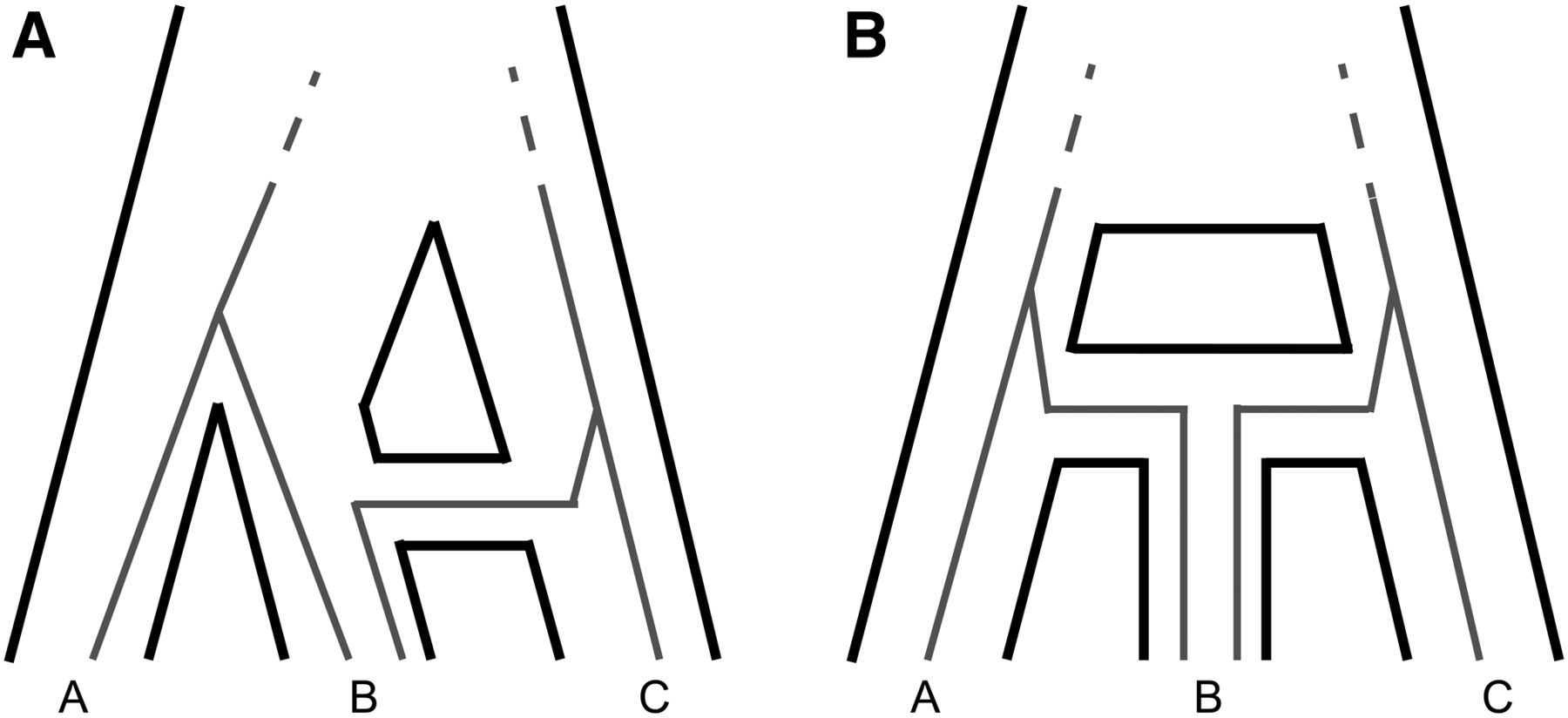
Genomic data has revealed that introgression, the hybridization and subsequent back-crossing of closely related species or isolated lineages, is common across the tree of life. I have been working on ways to take advantage of this abundance of data to make more detailed and biologically informed inferences about the occurrence, direction, timing, and biological factors affecting introgression.
Accounting for gene tree discordance in phylogenetic comparative methods
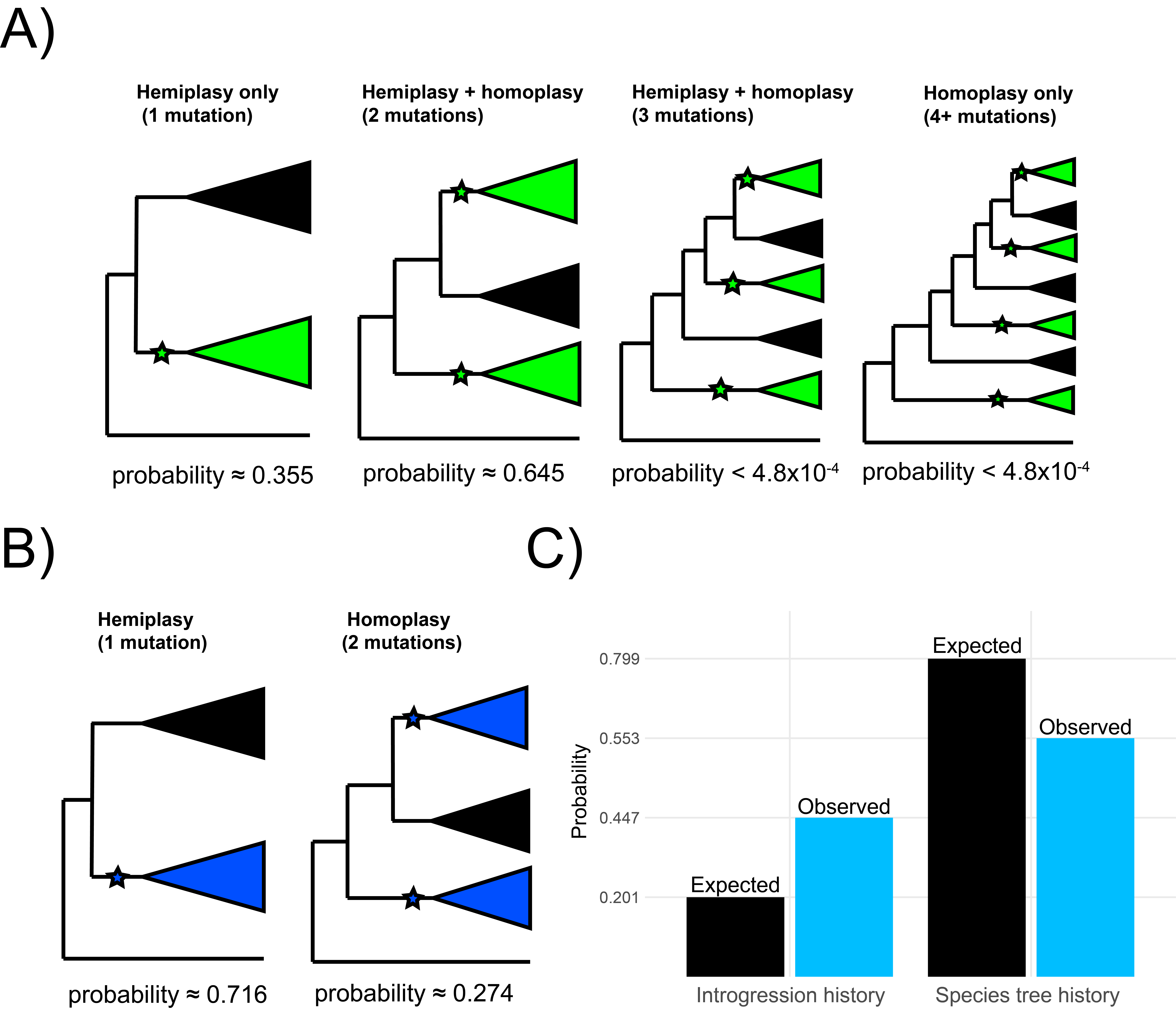
Gene tree discordance is a widespread phenomenon where the topology of trees inferred at individual loci disagree both with each other and the species tree topology. Two major causes are incomplete lineage sorting and introgression. Evolutionary changes along discordant gene tree branches results in potentially misleading patterns of trait evolution. Classic phylogenetic comparative methods do not account for these processes when making inferences about trait evolution. I have been developing theory and software with the goal of making more robust inferences in the presence of gene tree discordance.
Understanding macroevolutionary drivers of genome and transcriptome evolution
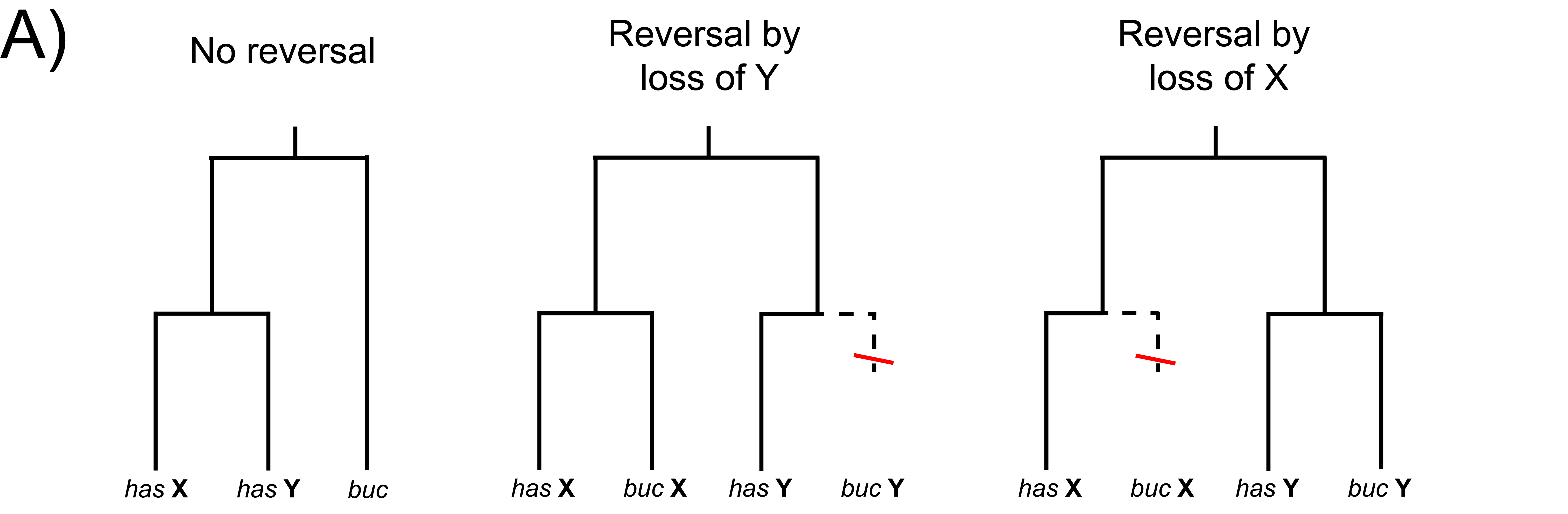
Genomes and transcriptomes vary widely among species. The forces shaping this variation are complex and, in many cases, not well understood. I am using modern genomic techniques to answer this question in a variety of systems, focusing on two main topics: 1) the evolution of transcriptional regulation associated with parallel phenotypic evolution; 2) the evolution of sex chromosome gain, loss, and turnover.